Regulation of Transcription
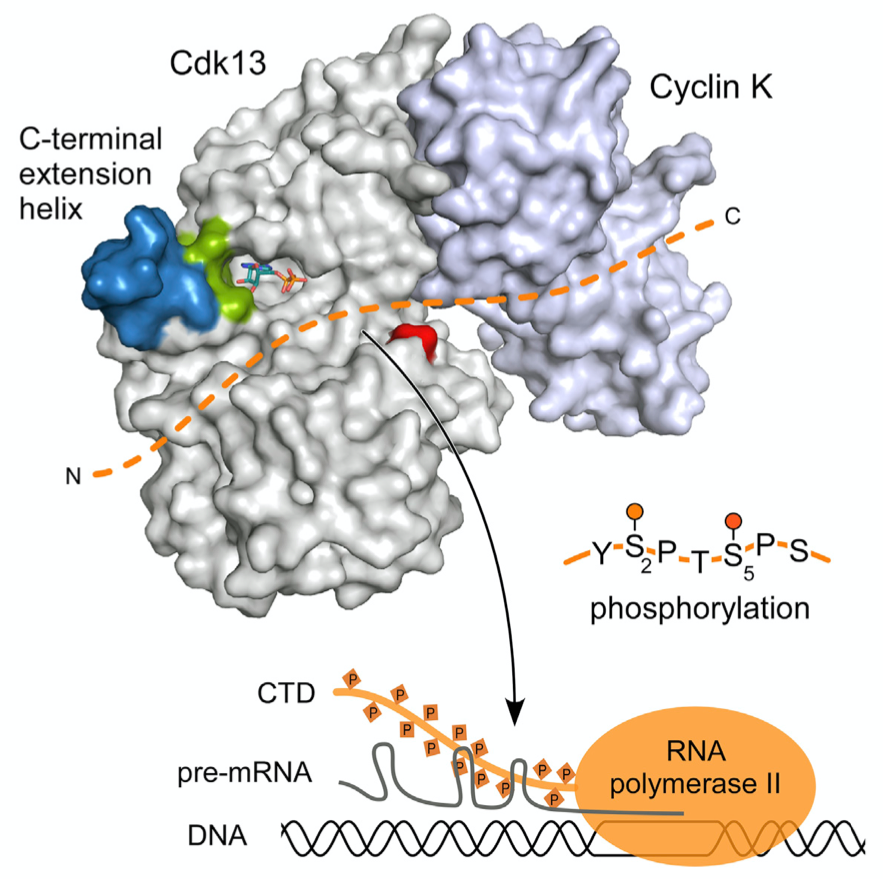
The transcription cycle is regulated by Cyclin-dependent kinases that phosphorylate the RNA polymerase II and general transcription factors. We analyse how the transition from transcription initiation to productive elongation, polyadenylation and termination is mediated in eukaryotic cells. After transcription initiation, the RNA polymerase II (RNAPII) pauses approximately 50-150 nucleotides downstream of the transcription start site. Release from this block requires the positive transcription elongation factor P-TEFb, which is a heterodimer composed of the cyclin-dependent kinase Cdk9 and the regulatory subunit Cyclin T. Cdk9 mediates the transition from transcription initiation to productive elongation of pre-mRNA transcripts by phosphorylation of the C-terminal domain (CTD) of RNAPII. Other cyclin-dependent kinases, Cdk7, CDK8, Cdk11, Cdk12 and Cdk13, contribute to transcriptional initiation, pre-mRNA processing and splicing regulation. We aim at analyzing the molecular and structural mechanisms that determine the activity and regulation of the transcription cycle controlling kinases. The malfunction of gene expression at a transcriptional level leads to a variety of diseases as multiple forms of cancer, leukaemia, HIV infection, and myocardial hypertrophy.
Selected reading
- Düster, R., et al. (2024) Structural basis of Cdk7 activation by dual T-loop phosphorylation. Nat. Commun. 15:6597.
- Insco, M.L., et al. (2023). Oncogenic CDK13 mutations impede nuclear RNA surveillance. Science 380:eabn7625.
- Düster, R., et al. (2022). Functional characterization of the human Cdk10/Cyclin Q complex. Open Biol. 12:210381.
- Kaltheuner, I.H., et al. (2021). Abemaciclib is a potent inhibitor of DYRK1A and HIP kinases involved in transcriptional regulation. Nat. Commun. 12:6607.
- Mayor-Ruiz, C. et al. (2020). Rational discovery of molecular glue degraders via scalablechemical profiling. Nat. Chem. Biol. 16, 1199-1207
- Olson, C.M., et al. (2019). Development of a selective CDK7 covalent inhibitor reveals predominant cell-cycle phenotype. Cell Chem. Biol.26, 792–803.
- Bugai, A., et al. (2019). P-TEFb activation by RBM7 shapes a pro-survival transcriptional response to genotoxic stress. Mol. Cell 74, 254–267.
- Krajewska, M., et al. (2019). CDK12 loss in cancer cells affects DNA damage response genes through premature cleavage and polyadenylation. Nat. Commun. 10:1757.
- Zhang, T., et al. (2016). Covalent targeting of remote cysteine residues to develop CDK12 and CDK13 inhibitors. Nat. Chem. Biol. 12, 876-884.
- Greifenberg, A.K., et al. (2016). Structural and functional analysis of the Cdk13/Cyclin K complex. Cell Rep. 14, 320–331.
- Bösken, C.A., et al. (2014). The structure and substrate specificity of human Cdk12/Cyclin K. Nat. Commun. 5:3505.
- Eick, D. and Geyer, M. (2013). The RNA polymerase II carboxy-terminal domain (CTD) code. Chem. Rev.113, 8456–8490.
- Czudnochowski, N., et al. (2012). Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition. Nat. Commun. 3:842.


