Teaching
At the Institute of Structural Biology, we teach advanced courses in protein biochemistry and structural biology for Master student classes and PhD courses. We are integrated in the Excellence Cluster 'ImmunoSensation2: the immunosensory system' at the University of Bonn and participate in the Master Science course 'Medical Immunosciences and Infection' as well as 'Chemistry'. In the following we give a short description of the key topics.
Structural biology
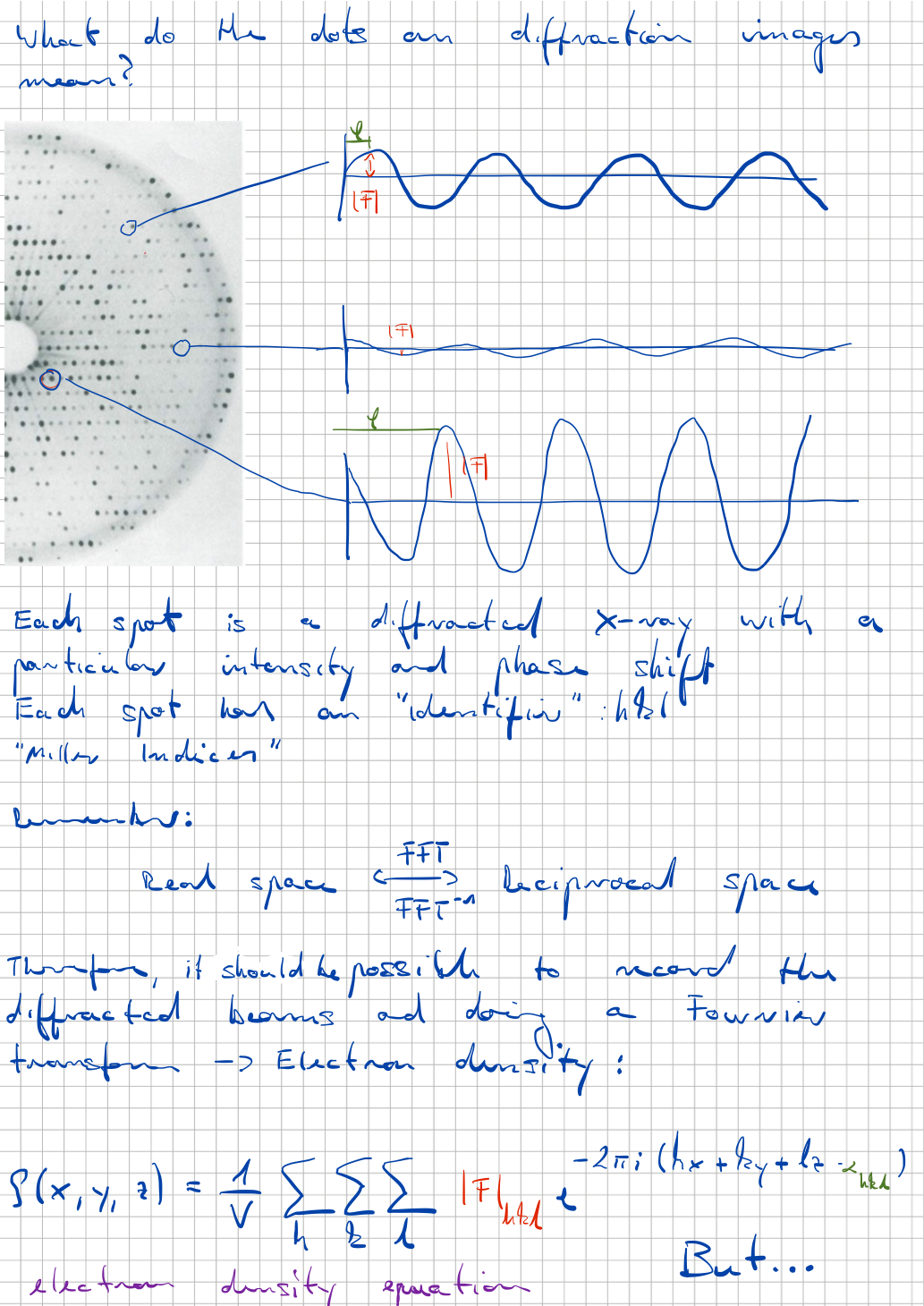
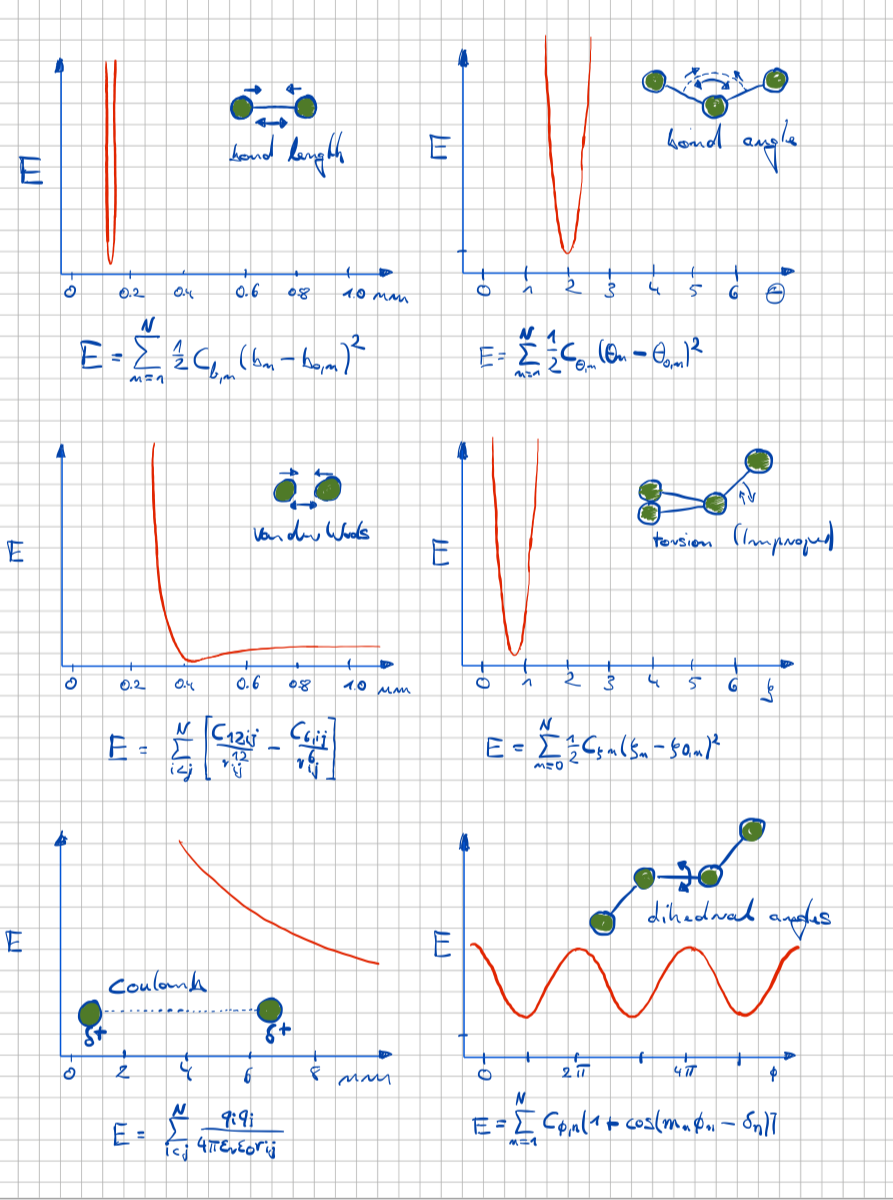
In this lecture series we teach the basic knowledge of the three main methods used to determine the three-dimensional structures of bio-molecules: X-ray crystallography, nuclear magnetic resonance (NMR) spectroscopy, and electron microscopy (EM). Students learn the requirements for the successful application of each technique, and the advantages and caveats of each particular method. Moreover, the skills to critically review the structure evaluation and description in scientific publications are taught.
Protein crystallography
The participants of this course will get an overview of the individual working steps for the structure determination of proteins by single crystal X-ray diffraction. Different techniques of protein crystallization, data collection and evaluation as well as structure determination will be learned. We also touch the history of structural biology, starting with the famous Photo 51 from Rosalind Franklin’s and Raymond Goslings's work and the interpretation by James Watson and Francis Crick, to restrained molecular dynamics calculations and modern 3D reconstructions from electron microscopy images.
Protein biochemistry
In this course the participants will get an overview of the major expression and purification techniques to gain recombinant protein. Starting with cloning and the design of expression vectors, we discuss how to determine domain boundaries and expression constructs. Various bioinformatic tools including codon optimization, secondary structure prediction or the determination of intrinsically unstructured regions will be considered for the design of protein expression constructs. Expression protocols for E.coli bacterial cells, baculo-virus infected insect cells, and eukaryotic cells in suspension culture will be discussed. State of the art affinity purification, gel filtration and fractionation are conveyed as well as analytical techniques for the protein characterization as size exclusion chromatography, dynamic light scattering, multi-angle light scattering and thermal denaturation. Students shall be trained to be able to design, express and purify proteins for further characterization.
If you are interested in doing an internship, a lab rotation, or your Bachelor or Master thesis in our labs, please contact Matthias Geyer or Gregor Hagelüken.
Structural biology
In this lecture series we teach the basic knowledge of the three main methods used to determine the three-dimensional structures of bio-molecules: X-ray crystallography, nuclear magnetic resonance (NMR) spectroscopy, and electron microscopy (EM). Students learn the requirements for the successful application of each technique, and the advantages and caveats of each particular method. Moreover, the skills to critically review the structure evaluation and description in scientific publications are taught.
Protein crystallography
The participants of this course will get an overview of the individual working steps for the structure determination of proteins by single crystal X-ray diffraction. Different techniques of protein crystallization, data collection and evaluation as well as structure determination will be learned. We also touch the history of structural biology, starting with the famous Photo 51 from Rosalind Franklin’s and Raymond Goslings's work and the interpretation by James Watson and Francis Crick, to restrained molecular dynamics calculations and modern 3D reconstructions from electron microscopy images.
Protein biochemistry
In this course the participants will get an overview of the major expression and purification techniques to gain recombinant protein. Starting with cloning and the design of expression vectors, we discuss how to determine domain boundaries and expression constructs. Various bioinformatic tools including codon optimization, secondary structure prediction or the determination of intrinsically unstructured regions will be considered for the design of protein expression constructs. Expression protocols for E.coli bacterial cells, baculo-virus infected insect cells, and eukaryotic cells in suspension culture will be discussed. State of the art affinity purification, gel filtration and fractionation are conveyed as well as analytical techniques for the protein characterization as size exclusion chromatography, dynamic light scattering, multi-angle light scattering and thermal denaturation. Students shall be trained to be able to design, express and purify proteins for further characterization.
If you are interested in doing an internship, a lab rotation, or your Bachelor or Master thesis in our labs, please contact Matthias Geyer or Gregor Hagelüken.